node_chain¶
Module: missions.operations.node_chain¶
Interface to node_chain using the BenchmarkNodeChain
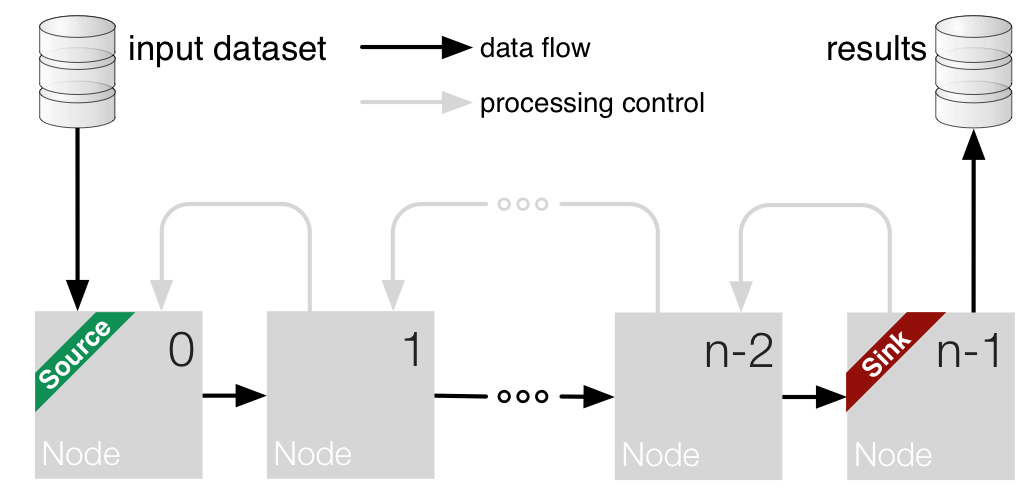
A process consists of applying the given node_chain on an input for a certain parameters setting.
Factory methods are provided that create this operation based on a specification in one or more YAML files.
Note
Complete lists of available nodes and namings can be found at: List of all Nodes.
See also
Specification file Parameters¶
type¶
Specifies which operation is used. For this operation you have to use node_chain.
(obligatory, node_chain)
input_path¶
Path name relative to the storage in your configuration file.
The input has to fulfill certain rules,
specified in the fitting: dataset_defs subpackage.
For each dataset in the input and each parameter combination
you get an own result.
(obligatory)
templates¶
List of node chain file templates which shall be evaluated.
The final node chain is constructed, by substituting specified parameters
of the operation spec file. Therefore you should use the format
__PARAMETER__. Already fixed and usable keywords are
__INPUT_DATASET__ and __RESULT_DIRECTORY__.
The spec file name may include a path and it is searched relative
the folder node_chains in the specs folder,
given in your configuration file you used, when starting pySPACE
(pySPACE.run.launch).
If no template is given, the software tries to use the parameters node_chain, which simply includes the node_chain description.
(recommended, alternative: `node_chain`)
node_chain¶
Instead of using a template you can use your single node chain directly in the operation spec.
(optional, default: `templates`)
runs¶
Number of repetitions for each running process. The run number is given to the process, to use it for choosing random components, especially when using cross-validation.
(optional, default: 1)
parameter_ranges¶
Dictionary with parameter names as keys and a list of values it should be replaced with. Finally a grid of all possible combinations is build and afterwards only those remain fulfilling the constraints.
(mandatory if parameter_setting is not used)
parameter_setting¶
If you do not use the parameter_ranges, this is a list of dictionaries giving the parameter combinations, you want to test the operation on.
(mandatory if parameter_ranges is not used)
constraints¶
List of strings, where the place holders for the parameter values are replaced as in the node chain template. After creating all possible combinations of parameters, they are inserted into this string and the string is evaluated like a Python expression. If it is not evaluated to True, the parameter is rejected. The test is performed for each constraint (string). For being used later on, a parameter setting has to pass all tests. Example:
__alg__ in ['A','B','C'] and __par__ == 0.1 or __alg__ == 'D'
(optional, default: [])
old_parameter_constraints¶
Same as constraints, but here parameters of earlier operation calls, e.g. windowing, can be used in the constraint def.
hide_parameters¶
Normally each parameter is added in the format {__PARAMETER__: value} added to the __RESULT_DIRECTORY__. Every parameter specified in the list hide_parameters is not added to the folder name. Be careful not to have the same name for different results.
(optional, default: [])
storage_format¶
Some datasets give the opportunity to choose between different
formats for storing the result. The choice can be specified here.
For details look at the dataset_defs documentation.
If the format is not specified, the default of the
dataset is used.
(optional, default: default of dataset)
store_node_chain¶
If True, the total state of the node chain after the processing is stored to disk. This is done separately for each split and run. If store_node_chain is a tuple of length 2—lets say (i1,i2)—only the subflow starting at the i1-th node and ending at the (i2-1)-th node is stored. This may be useful when the stored flow should be used in an ensemble. If store_node_chain is a list with indices, all nodes in the chain with the corresponding indices are stored. This might be useful to exclude specific nodes that do not change the data dimension / type, like e.g. visualization nodes.
Note
Since yaml cannot recognize tuples, string parameters are evaluated before type testing (if boolean, tuple, list) is done.
(optional, default: False)
compression¶
If your result is a classification summary,
all the created sub-folders are zipped with the zipfile module,
since they are normally not needed anymore,
but you may have numerous folders, making coping difficult.
To switch this of, use the value False and if you want no
compression, use 1. If all the sub-folders (except a single one)
should be deleted, set compression to delete.
(optional, default: 8)
Exemplary Call¶
type: node_chain
input_path: "my_data"
runs : 2
templates : ["example_node_chain1.yaml","example_node_chain2.yaml","example_node_chain3.yaml"]
type: node_chain
input_path: "my_data"
runs : 2
parameter_ranges :
__prob__ : eval(range(1,4))
node_chain :
-
node: FeatureVectorSource
-
node: CrossValidationSplitter
parameters:
splits : 5
-
node: GaussianFeatureNormalization
-
node: LinearDiscriminantAnalysisClassifier
parameters:
class_labels: ["Standard","Target"]
prior_probability : [1,__prob__]
-
node: ThresholdOptimization
parameters:
metric : Balanced_accuracy
-
node: Classification_Performance_Sink
parameters:
ir_class: "Target"
type: node_chain
input_path: "example_data"
templates : ["example_flow.yaml"]
backend: "local"
parameter_ranges :
__LOWER_CUTOFF__ : [0.1, 1.0, 2.0]
__UPPER_CUTOFF__ : [2.0, 4.0]
constraints:
- "__LOWER_CUTOFF__ < __UPPER_CUTOFF__"
runs : 10
type: node_chain
input_path: "my_data"
runs : 2
templates : ["example_node_chain.yaml"]
parameter_setting :
-
__prob__ : 1
-
__prob__ : 2
-
__prob__ : 3
Here example_node_chain.yaml is defined in the node chain specification folder as
-
node: FeatureVectorSource
-
node: CrossValidationSplitter
parameters:
splits : 5
-
node: GaussianFeatureNormalization
-
node: LinearDiscriminantAnalysisClassifier
parameters:
class_labels: ["Standard","Target"]
prior_probability : [1,__prob__]
-
node: ThresholdOptimization
parameters:
metric : Balanced_accuracy
-
node: Classification_Performance_Sink
parameters:
ir_class: "Target"
Inheritance diagram for pySPACE.missions.operations.node_chain:

Class Summary¶
NodeChainOperation(processes, ...[, ...]) |
Load configuration file, create processes and consolidate results |
NodeChainProcess(node_chain_spec, ...[, ...]) |
Run a specific signal processing chain with specific parameters and set and store results |
Classes¶
NodeChainOperation¶
-
class
pySPACE.missions.operations.node_chain.NodeChainOperation(processes, operation_spec, result_directory, number_processes, create_process=None)[source]¶ Bases:
pySPACE.missions.operations.base.OperationLoad configuration file, create processes and consolidate results
The operation consists of a set of processes. Each of these processes consists of applying a given
BenchmarkNodeChainon an input for a certain configuration of parameters.The results of this operation are collected using the consolidate method that produces a consistent representation of the result. Especially it collects not only the data, but also saves information of the in and out coming files and the used specification files.
Class Components Summary
_createProcesses(processes, ...)_get_result_dataset_dir(base_dir, ...)Determines the name of the result directory consolidate([_])Consolidates the results obtained by the single processes into a consistent structure of collections that are stored on the file system. create(operation_spec, result_directory[, ...])A factory method that creates the processes which form an operation based on the information given in the operation specification, operation_spec. -
__init__(processes, operation_spec, result_directory, number_processes, create_process=None)[source]¶
-
classmethod
create(operation_spec, result_directory, debug=False, input_paths=[])[source]¶ A factory method that creates the processes which form an operation based on the information given in the operation specification, operation_spec.
In debug mode this is done in serial. In the other default mode, at the moment 4 processes are created in parallel and can be immediately executed. So generation of processes and execution are made in parallel. This kind of process creation is done independently from the backend.
For huge parameter spaces this is necessary! Otherwise numerous processes are created and corresponding data is loaded but the concept of spreading the computation to different processors can not really be used, because process creation is blocking only one processor and memory space, but nothing more is done, till the processes are all created.
-
classmethod
_createProcesses(processes, result_directory, operation_spec, parameter_settings, input_collections)[source]¶
-
NodeChainProcess¶
-
class
pySPACE.missions.operations.node_chain.NodeChainProcess(node_chain_spec, parameter_setting, rel_dataset_dir, run, split, storage_format, result_dataset_directory, store_node_chain=False, hide_parameters=[])[source]¶ Bases:
pySPACE.missions.operations.base.ProcessRun a specific signal processing chain with specific parameters and set and store results
This is an atomic task,which takes a dataset (EEG-data, time series data, feature vector data in different formats) as input and produces a certain output (e.g. ARFF, pickle files, performance tabular). The execution of this process consists of applying a given NodeChain on an input for a certain configuration.
- Parameters
node_chain_spec: String in YAML syntax of the node chain. parameter_setting: parameters chosen for this process rel_dataset_dir: location of input file relative to the pySPACE storage, specified in the configuration file run: When process is started repeatedly, to deal with random components, we have to forward the current run number to these components. split: When there are different splits, forward the split number. storage_format: Choice of format, the resulting data is saved. This should always fit to the sink node specified as the last node in the chain. result_dataset_directory: Folder where the result is saved at. store_node_chain: option to save as pickle file the total state of the node chain after the processing; separately for each split in cross validation if existing
Class Components Summary
__call__()Executes this process on the respective modality _check_node_chain_dataset_consistency(...)Checks that the given node_chain can process the given dataset
